Token for Submission
If you don’t receive the submission token within 3 days after registration, please check your email’s spam folder first. Feel free to contact us if you have any questions.
Overview
The ICTS Challenge 2025 invites participants to advance the field of brain tumor segmentation, a critical component in the diagnosis and treatment of brain tumors. This year, we introduce a unique twist: the use of synthetic training datasets. Your challenge is to develop effective segmentation models that perform well on real-world clinical data, despite being trained on synthetic images.
Leaderboard
Background
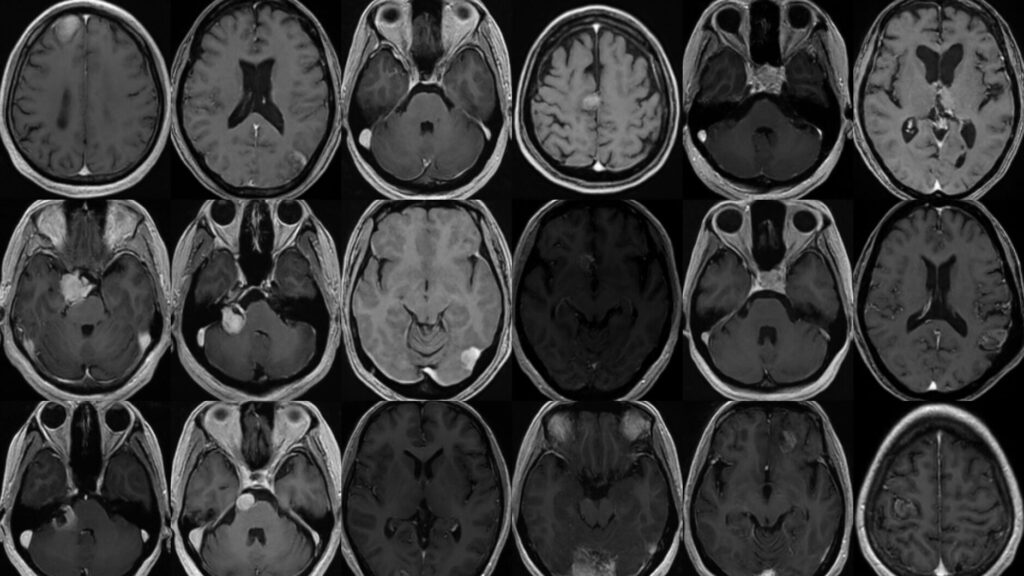
Brain tumor segmentation plays a crucial role in patient diagnosis and treatment. However, most deep learning methods rely on pre-processed images, limiting their applicability in real clinical settings. Recognizing this gap, the ICTS Challenge 2025 focuses on unnormalized clinical images and a wider range of brain tumors, including but not limited to brain metastases, meningiomas, schwannomas, and pituitary adenomas. These conditions present a significant challenge due to their varying sizes and complexities in imaging representation.
Task
Register your team officially using the registration form. You can join the challenge either as a team or as an individual participant. Your task is to create a model that can accurately identify brain tumors in real patient data. Although you will train your model using synthetic images, its performance will be evaluated using actual clinical data. You will need to present your work during the workshop on December 26, 2025.
Important Dates
| 2025-7-20 | Release of training data – 1 | Med-DDPM v1.0 |
| 2025-7-31 | Release of training data – 2 | Med-DDPM v2.0 |
2025-12-16 Updated | Release of evaluation script and sample submission script | public.zip Updated |
| 2025-12-13 | Release of test image information (dimensions, spacing, scalar range) | image_info.txt |
| 2025-12-24 23:59 (GMT+8) | Freeze Leaderboard | Submissions are still possible, but the results will be hidden from others. |
| 2025-12-25 23:59 (GMT+8) | Deadline for model submission | |
| 2025-12-26 10:00-12:00 (GMT+8) | Presentations at workshop (Hybrid) | National Taiwan University Hospital Clinical Research Building 台大醫院臨床研究大樓9F大會議室 |
Prizes
Sponsored by Taiwan Kuangli Co., Ltd. 台灣光麗實業


Winners are determined based on the final evaluation scores, with awards presented to the top three teams. Note: Awards are contingent upon presenting at our workshop in Taipei on December 26, 2025.
Dataset and Evaluation
In compliance with strict patient privacy and health data regulations, we are unable to share real patient-derived datasets openly. Instead, we offer a viable solution using synthetic datasets [1], eliminating the need for data anonymization, image cropping, or feature exclusion. Our challenge provides an extensive synthetic training dataset, generated using the Med-DDPM model [2], featuring contrast-enhanced T1-weighted images paired with tumor labels. Additionally, we supply the source code and pretrained weights of the Med-DDPM model [3], empowering participants to create more data by customizing input masks.
The final model evaluation will utilize real patient images. Winners will be determined based on the highest Dice similarity coefficient score achieved during this evaluation phase.
Important Points for Participants:
The ICTs challenge is focused on training an accurate tumor segmentation model for brain T1c MRI images. Participants should pay attention to the following details:
The principle of “Garbage in, garbage out” highlights the importance of data preprocessing as a first step. Mask images include labels 0, 1, and 2: 0 represents the background, 1 the brain area, and 2 the tumor area. Participants are required to submit models that predict only the tumor mask (label 1), meaning the model should identify tumor pixels as label 1. When training the segmentation model, participants must preprocess the dataset and modify mask labels appropriately. Specifically, labels in the first dataset should be changed from brain label 1 to 0 and tumor label 2 to 1.
We have made our pretrained image synthesis model, Med-DDPM, publicly available for participants to generate additional data for . To create synthetic images, this model requires a mask image with three labels: 0 for the background, 1 for the brain area, and 2 for the tumor area. Therefore, participants can use the dataset’s mask images for this purpose. Participants can manipulate these mask images to create new masks, which can then be fed into Med-DDPM to generate new images.
If you have any questions or require clarifications regarding this challenge, please feel free to contact us at mobaidoctor@gmail.com at any time. Thank you.
References
[1] Strickland, E. (2022, March 10). Are you still using real data to train your AI? IEEE Spectrum. Retrieved July 10, 2022, from https://spectrum.ieee.org/synthetic-data-ai
[2] Z. Dorjsembe, H. K. Pao, S. Odonchimed, and F. Xiao, “Conditional diffusion models for semantic 3D brain MRI synthesis,” IEEE Journal of Biomedical and Health Informatics, vol. 28, no. 7, pp. 4084-4093, Jul. 2024, doi: 10.1109/JBHI.2024.3385504.
[3] “Med-DDPM: Medical Denoising Diffusion Probabilistic Models,” GitHub repository, 2023. [Online]. Available: https://github.com/mobaidoctor/med-ddpm/
About US
Host: Institute of Medical Device and Imaging ,National Taiwan University
Organizer: Intelligent Medical Device Laboratory, National Taiwan University
Sponsors: National Science and Technology Council, Taiwan Kuangli Co., Ltd.